SDL Digital Patient
A cooperation of TU Darmstadt
and RWTH Aachen University
and RWTH Aachen University
Simulation and Data Lab
The SDL Digital Patient aims to support the implementation of a digital counterpart to the patient in clinic, in order to simulate and predict the outcome of disease and therapy.
Our target audience consists of clinicians and researchers in biomedical research with suitable data but not necessarily the skillset to run simulations in HPC environments.
We offer a diverse set of methods ranging from data science and machine learning (in particular image analysis), to molecular dynamics simulations and hybrid modelling of patient-scale models.
If you have questions for other groups or general questions like access to the HPC infrastructure, have a look at our support website.
Current research topics:
- Digital Patient ‘Oncology’ – collaboration with Prof. Tim Brümmendorf & Prof. Steffen Koschmieder (Uniklinik Aachen)
- Digital Patient ‘Pain’ – collaboration with Prof. Angelika Lampert (Uniklinik Aachen)
Support activities:
- Workshops with interested researchers to establish Digital Patient ‘Demonstrators’ that serve as exemplar for future implementations in other disease contexts
- Curation of a biomedical data ‘meta’ repository that includes a list of suitable data and instructions how to access these (restricted access)
- Consulting service to assist with HPC implementation
Gallery
Project partners
Members
Publications
2024
- How exascale computing can shape drug design: A perspective from multiscale QM/MM molecular dynamics simulations and machine learning-aided enhanced sampling algorithms (G. Rossetti and D. Mandelli), Current Opinion in Structural Biology
- Cholesterol modifies Nav1.7 – an in-silico and in-vitro analysis (S. Albani, V.S.B. Eswaran, A. Piergentili, P.C.T. de Souza, A. Lampert, G. Rossetti), submitted to Nature Communications Biology
- Hyperexcitability of stem cell-derived sensory neurons of erythromelalgia patient normalized by divalent ions (A.K. Kalia, A. Kelly, D. Tavares-Ferreira, M.A.S. Toledo, C. Rösseler, A. Piergentili, S. Albani, A. Kostritskii, P. Hautvast, A E. Pickering, J.P. Dunham, B.N.I. Kurth, A.J. Gaebler, J.P. Machtens, G. Rossetti, T.J. Price, M. Zenke, A. Lampert), Annal of Neurology, under review
2023
- Diagnostic Expert Advisor: A platform for developing machine learning models on medical time-series data (Richard Polzin, Sebastian Fritsch, Konstantin Sharafutdinov, Gernot Marx, Andreas Schuppert), SoftwareX
2022
- TransNorm: Transformer Provides a Strong Spatial Normalization Mechanism for a Deep Segmentation Model (Reza Azad, Mohammad T. Al-Antary, Moein Heidari, and Dorit Merhof), IEEE Access
- Instance Segmentation of Dense and Overlapping Objects via Layering (Long Chen, Yuli Wu and Dorit Merhof), The British Machine Vision Conference (BMVC 2022)
- HiFormer: Hierarchical Multi-scale Representations Using Transformers for Medical Image Segmentation (Moein Heidari, Amirhossein Kazerouni, Milad Soltany, Reza Azad, Ehsan Khodapanah Aghdam, Julien Cohen-Adad, Dorit Merhof), IEEE/CVF Winter Conference on Applications of Computer Vision (WACV2023)
- Enhanced Sampling Approach to the Induced-Fit Docking Problem in Protein–Ligand Binding: The Case of Mono-ADP-Ribosylation Hydrolase Inhibitors, (Riccardo Capelli, Paolo Carloni, Bernhard Lüscher, Jinyu Li, Giulia Rossetti), Journal of Chemical Theory and Computation
Conferences
- P. Gräbel, J. Thull, M. Crysandt, B. M. Klinkhammer, P. Boor, T. H. Brümmendorf, and D. Merhof, “Automatic embedding interventions for the classification of hematopoietic cells,” in 26th International Conference on Pattern Recognition (ICPR), 2022
- P. Gräbel, J. Thull, M. Crysandt, B. M. Klinkhammer, P. Boor, T. H. Brümmendorf, and D. Merhof, “Analysis of automatically generated embedding guides for cell classification,” in IEEE International Conference on Image Processing Theory and Tools and Applications (IPTA), 2022
- Qianqian Zhao, P. Gräbel, J. Thull, M. Crysandt, B. M. Klinkhammer, P. Boor, T. H. Brümmendorf, and D. Merhof, “Spatial maturity regression for the classification of hematopoietic cells,” in IEEE International Conference on Image Processing Theory and Tools and Applications (IPTA), 2022.

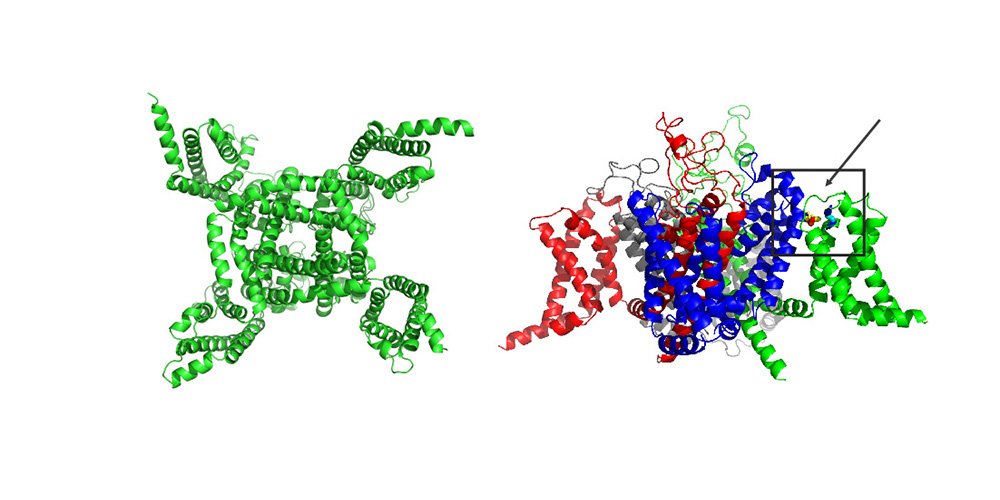 Protein structure of sodium channel Nav1.7 (© Alessia Piergentili/Giulia Rosetti)
Protein structure of sodium channel Nav1.7 (© Alessia Piergentili/Giulia Rosetti) 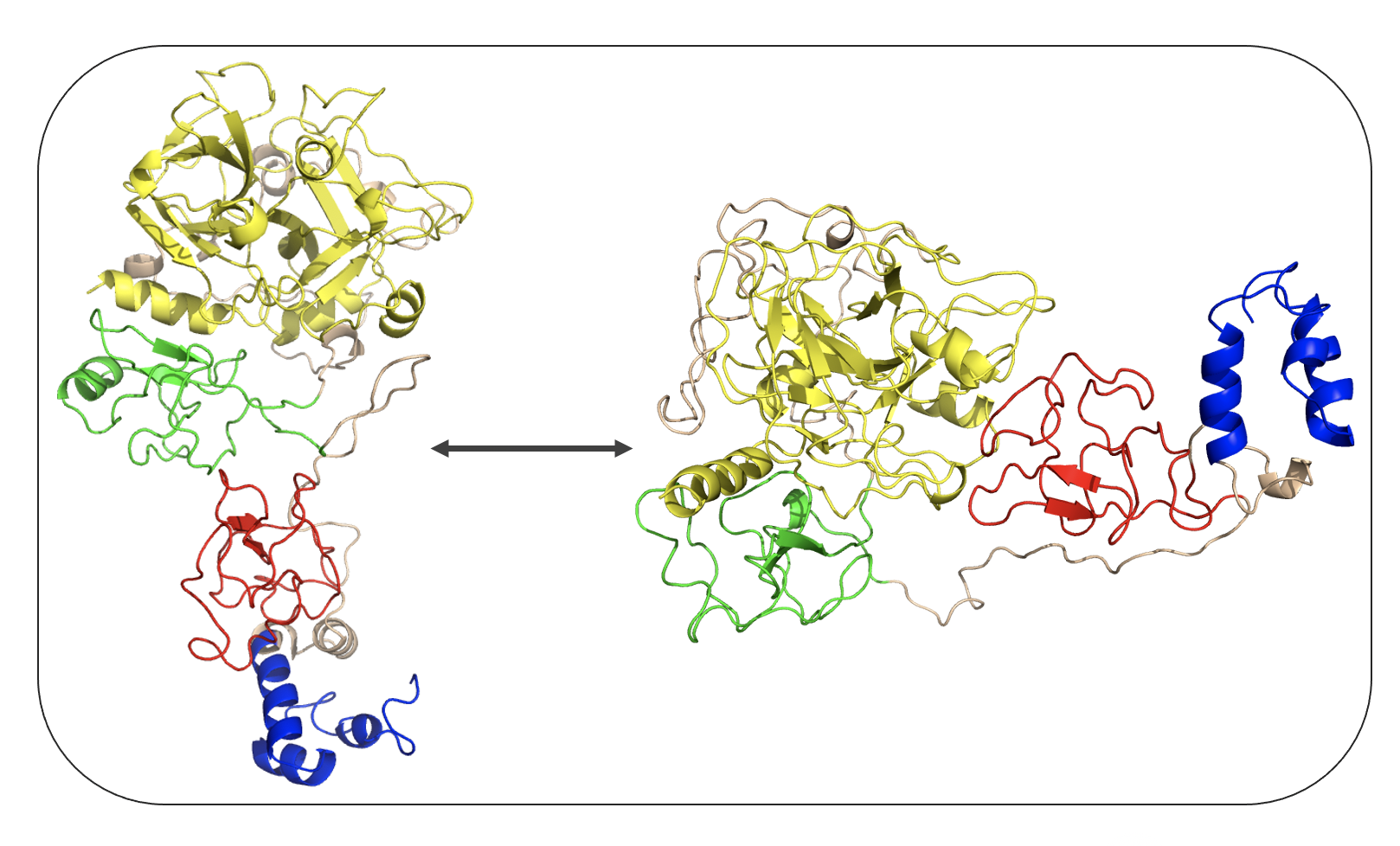 Protein structure of Prothrombin - open and closed conformations (© Alessia Piergentili/Giulia Rosetti)
Protein structure of Prothrombin - open and closed conformations (© Alessia Piergentili/Giulia Rosetti) 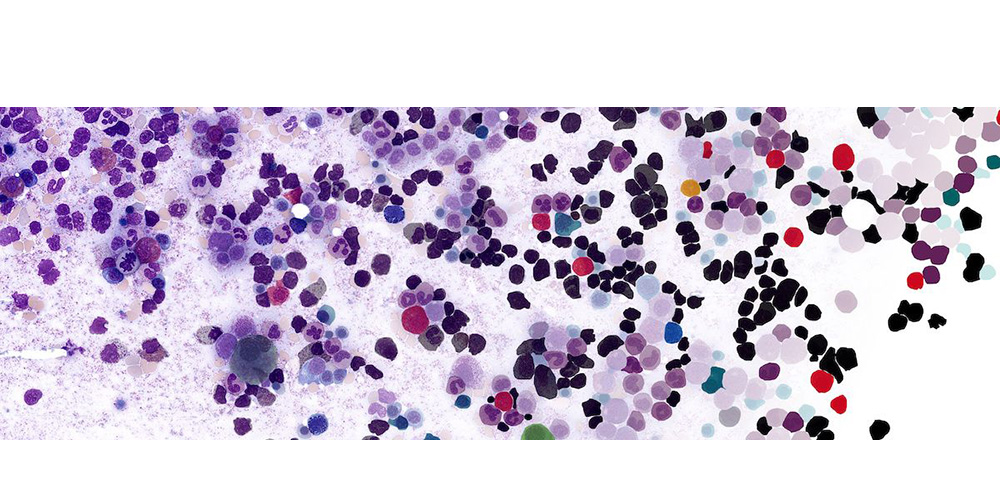 Automated image segmentation and annotation of bone marrow cell types (© Philip Gräbel and Dorit Merhoff)
Automated image segmentation and annotation of bone marrow cell types (© Philip Gräbel and Dorit Merhoff) 



