Project
Exploring the Free Energy Landscape of the Hcn Channels Voltage Sensing Domain Activation by Metadynamics
HCN channels are the most complex channels in the superfamily of voltage-gated ion channels in terms of their long activation time scale (milliseconds-seconds), unique structure and activation pathway, and the high number of degrees of freedom (tetrameric channels). Therefore, sampling the active state requires either performing very long molecular dynamics simulations or using advanced enhanced sampling techniques (e.g. metadynamics, On- the-fly Probability Enhanced Sampling (OPES) approaches, string methods based on swarms of trajectories). In this project, we adopted the second approach employing both the metadynamics method and the OPES technique.
Project Details
Project term
February 15, 2022–August 27, 2022
Affiliations
Forschungszentrum Jülich
RWTH Aachen University
Institute
Department of Biology
Principal Investigator
Methods
By exploiting the CLAIX-2018 supercomputer, we were able to get approximately a 30 ns/ day and 15 ns/day simulation rates when performing metadynamics and OPES simulations, respectively. Another difficulty in simulating our system is that there are in principle more than 20 degrees of freedom in each of the four subunits that would be important to include in the definition of the collective variable. This means that we needed also some advanced strategy to find or optimise the collective variable choices. For the metadynamics simulations we define a so-called pathway collective variable and for the OPES simulations we built a collective variable by exploiting the outcome from a deep learning algorithm. We had already collected two 400ns-long simulations at two different protonation states. Then, in this computational period we extended these plain MD simulations for an additional one microsecond and obtained stable structures (Fig. 1). We already had a test metadynamics simulation as well, which we used to initially explore the active state.
Results
The long MD trajectories obtained in the first part of the project were used to study the Hbond networks, which gave us meaningful insights to understand which key amino acids are responsible for pH sensitivity (Fig. 2). In the second part of the project, the metadynamics simulations were accomplished, and from them we reconstructed the free energy landscape reported in Fig. 3. These results were published in the final Ph.D thesis of Dr. Somayeh Asgharpour.
Discussion
Finally, in the last part of the project we performed the OPES simulations. The analysis of this part is still in progress, but the structure obtained through these simulations (Fig. 4) is very close to the available experimental active structure of the HCN2 channels (PDB id: 6uqf). All project results will be published after the analysis is completed.
Additional Project Information
DFG classification: 201-02 Biophysics
Cluster: CLAIX
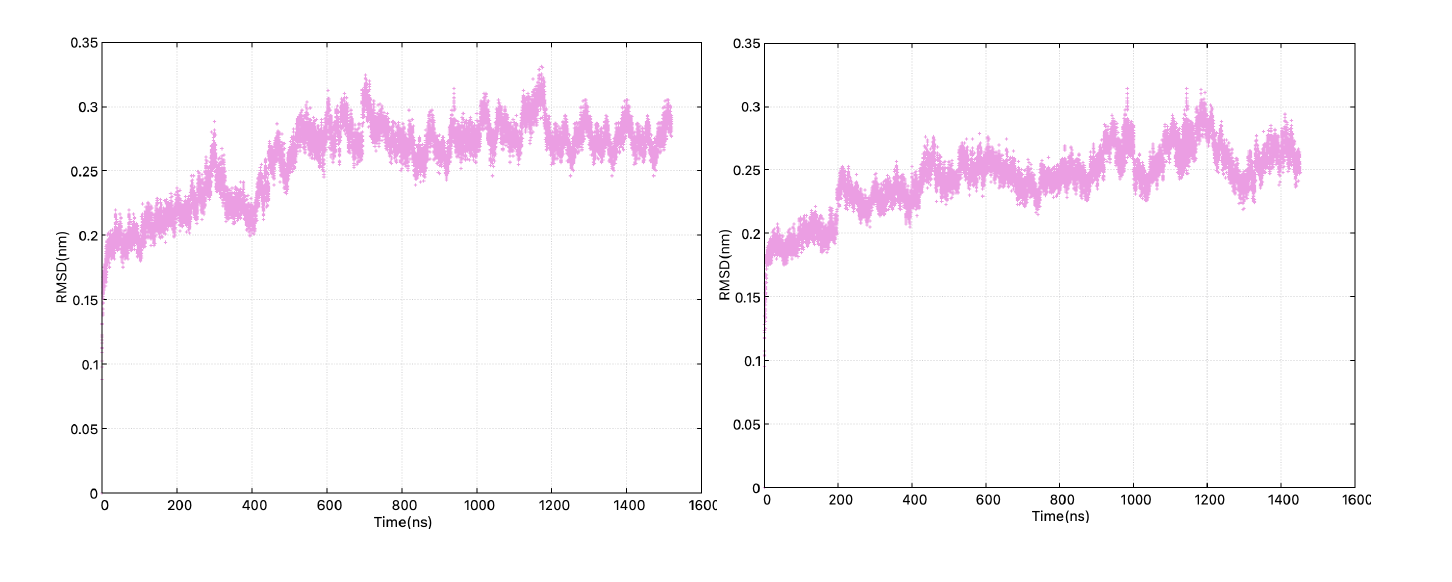 Figure 1, The RMSD of the transmembrane domain in the long MD simulation for systems.
Figure 1, The RMSD of the transmembrane domain in the long MD simulation for systems. 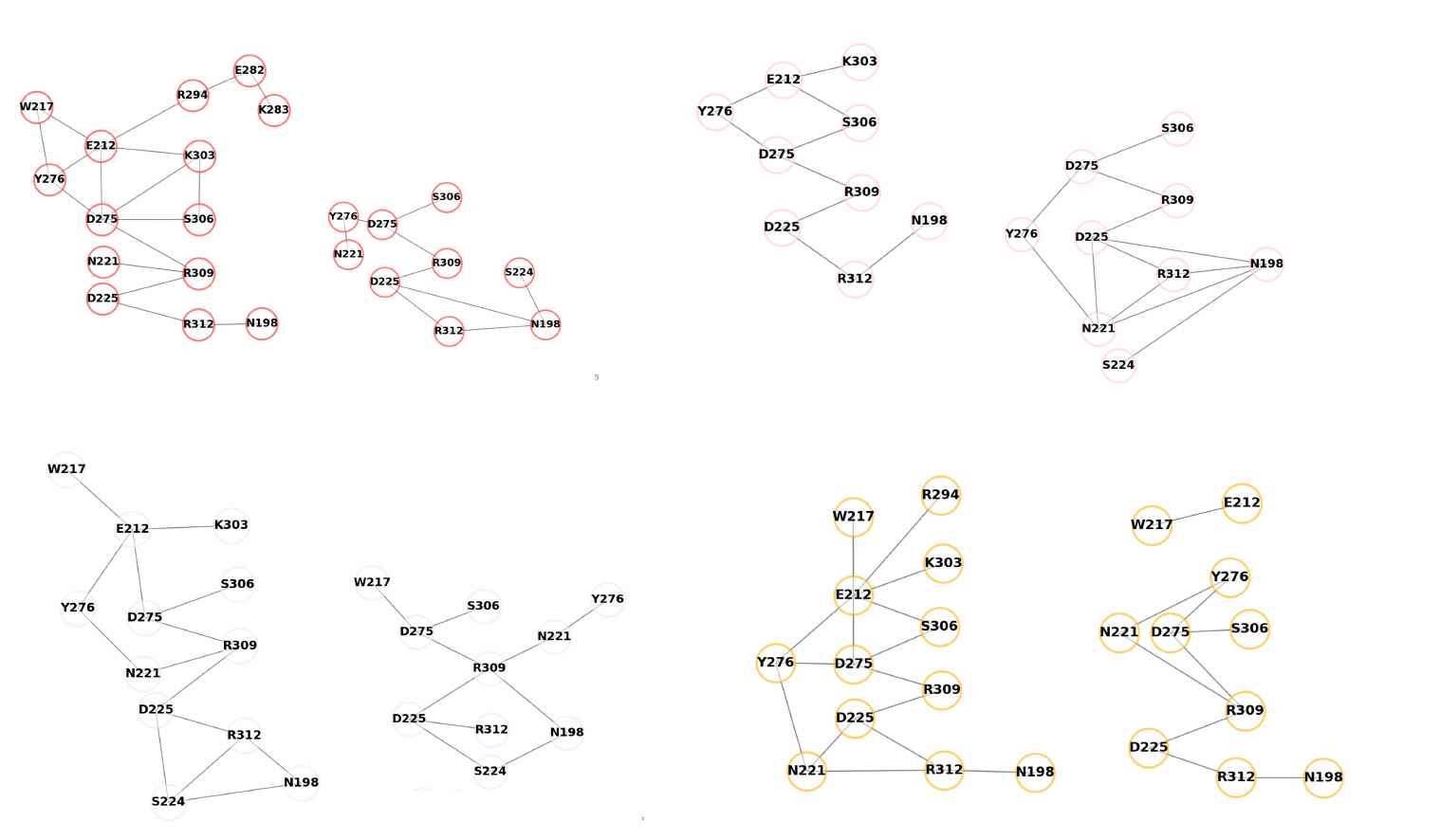 Figure 2, The changes in the H-bond network of the voltage sensing domain in all subunits upon the
protonation of GLU212 based on graph analysis of 1 microsecond long MD trajectories.
Figure 2, The changes in the H-bond network of the voltage sensing domain in all subunits upon the
protonation of GLU212 based on graph analysis of 1 microsecond long MD trajectories. 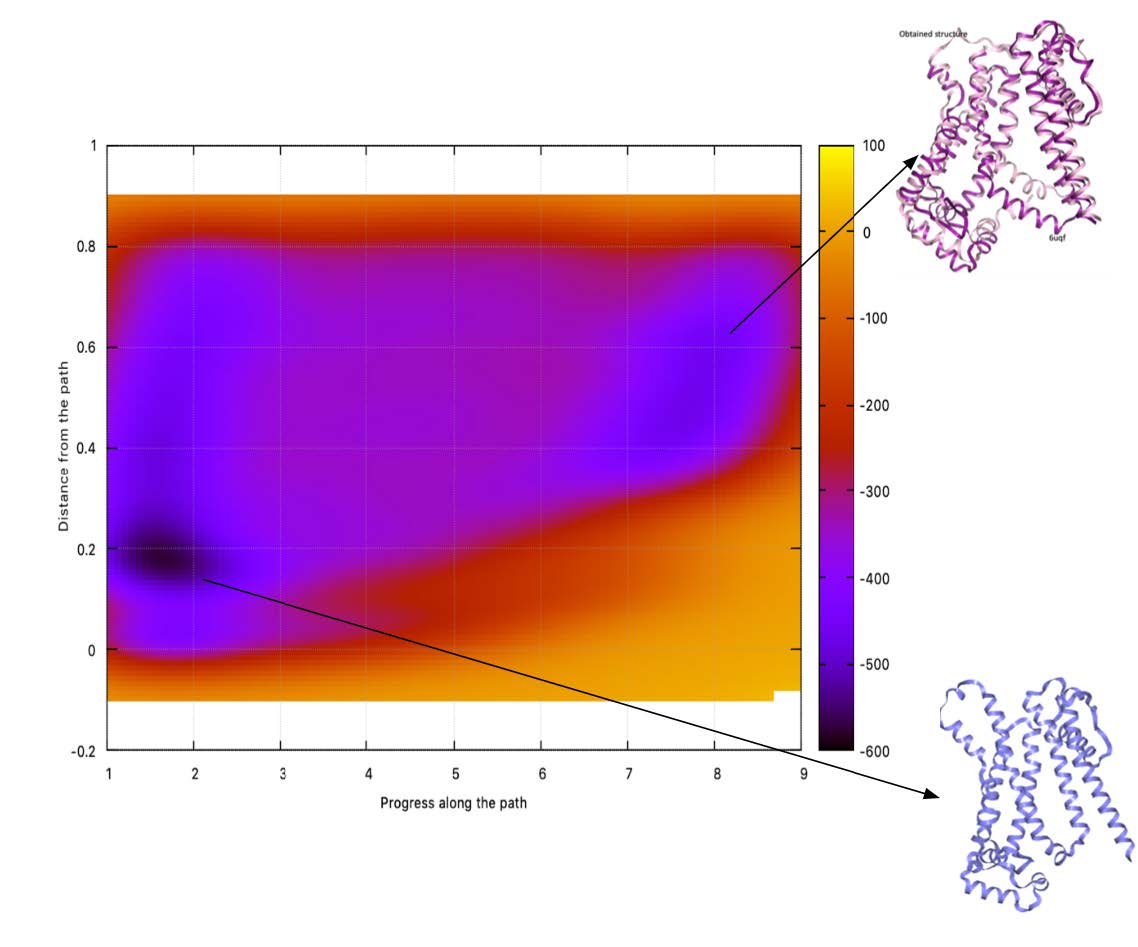 Figure 3, The free energy landscape of the activation of the HCN2 channel obtained by the pathway
metadynamics approach.
Figure 3, The free energy landscape of the activation of the HCN2 channel obtained by the pathway
metadynamics approach. 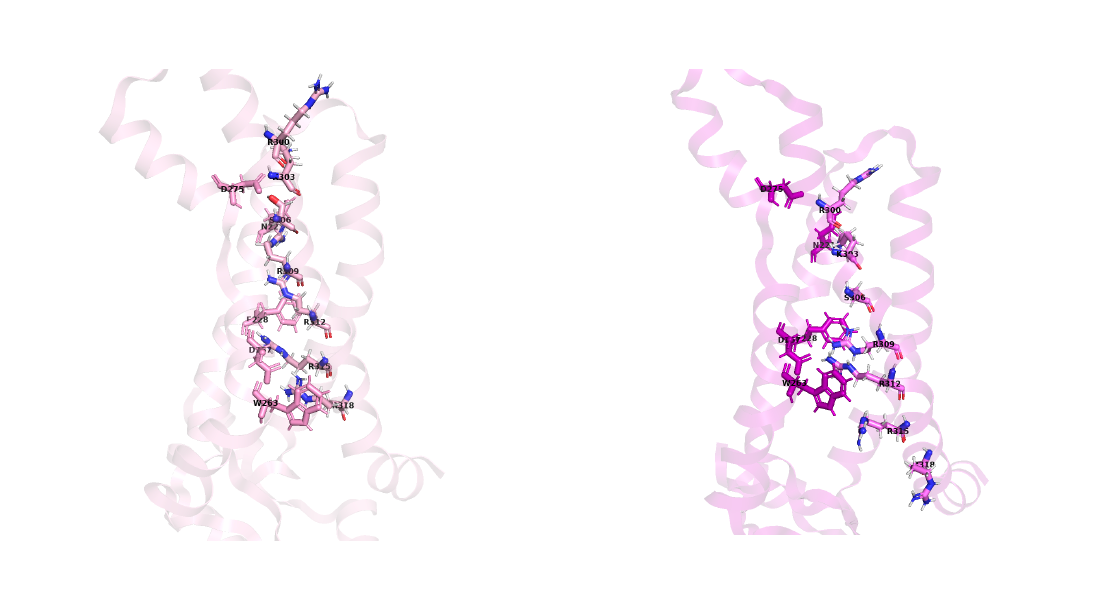 Figure 4, The structure obtained in OPES simulations and important motions.
Figure 4, The structure obtained in OPES simulations and important motions.