Project
Sodium Channel Nav1.7: Molecular Insights Into Erythromelalgia Mutation Q875E
This project is in collaboration with Prof. Dr. Angelika Lampert and her team from the Department of Neurophysiology at Uniklinik Aachen to investigate the neuronal phenotype of a patient carrying the Q875E mutation in the voltage-gated sodium channel Nav1.7. Navs are responsible for action potential and propagation in excitable cells. Within the 9 human Navs in the protein family, Nav1.7 is involved in pain mechanisms, especially in the development and transmission of inflammatory pain. Navs channelopathy are a group of disorders that cause a reduction of the channel fast inactivation or an enhancing of channel activation, both leading to a persistent sodium current. These include Primary Erythromelalgia, which has been shown to be caused by a so-called gain-of-function mutation of the channel, the Q875E mutation. A previous experimental study from Prof. Lampert’s lab suggested that this mutation stabilizes the activated state of the voltage sensing domain of the channel (VSDI) via the formation of a salt bridge between R214 on Domain I and E875 on Domain II, stabilizing the open conformation of the channel. The latter interaction can be reversed by increasing the concentration of divalent ions in solution. Given the potential of Mg2+ as a therapeutic agent for alleviating the symptoms in patients carrying this disease, our computational analysis aimed to elucidate the molecular mechanism underlying this potential treatment. To achieve this, we performed molecular dynamics (MD) simulations on the Nav1.7 on High-Performance Computing (HPC) CLAIX resources. The parallel computing capabilities of HPC were crucial in managing the computational demands of these simulations, enabling us to gain insights into the molecular basis of the proposed Mg2+ therapy.
Project Details
Project term
June 1, 2023–June 30, 2024
Affiliations
Forschungszentrum Jülich
Institute
Computational Biomedicine Institute
Project Manager
Principal Investigator
Methods
We employed the Martini 3.0.0 coarse-grained force field in conjunction with the GROMACS 2021.6 simulation software to model the Nav1.7 channel embedded in a neuronal membrane. The simulations were conducted for 4μs under varying concentrations of divalent cations in solution, allowing us to capture the system’s behavior over microsecondscale trajectories. MDAnalysis has been used to measure R214-Q875E distances residues.
Results
Two systems has been modeled starting from the most updated Nav1.7 cryo-EM structure showing the VSDI in the activated state and currently available in the Protein Data Bank at the beginning time of the project (PDB ID: 7W9K): the ion channel with the mutation (Q875E) and without (wild-type WT). The wild-type (WT) channel and the Q875E mutant were simulated under control (4 mM CaCl2) and increased (10 mM CaCl2) divalent ion concentrations, in line with experimental conditions. Our focus was on analyzing the interaction between the mutated residue (E875) and the gating charge residue (R214) in the first voltage sensing domain (VSD I). The negatively charged E875 in the pore domain is hypothesized to interact with R214 on VSD I, potentially stabilizing the active conformation of the latter. We observed that, at 4 mM CaCl2, the distance between E875 and R214 was shorter in the mutant compared to the WT channel. Interestingly, in the 10 mM CaCl2 simulations, the E875-R214 distance in the mutant increased, becoming comparable to that in the WT channel (Figure 1).
Discussion
Our molecular dynamics simulation studies indicate that the Q875E mutation indeed stabilizes VSD I in an active conformation through the formation of an electrostatic interaction with R214. This finding aligns with experimental evidence showing that increasing the concentration of divalent ions can mitigate the pathogenic effect of this mutation. Specifically, positive divalent ions appear to neutralize the local electrostatic influence of the mutated E875 residue, thereby inhibiting the E875-R214 interaction and disrupting the overly stabilized active conformation of VSD I. These findings support the potential of Mg2+ as a therapeutic option to alleviate pain symptoms in patients.
Additional Project Information
DFG classification: 201-02 Biophysics
Software: Gromacs
Cluster: CLAIX
Publications
Albani S. et al., Depletion of membrane cholesterol modifies structure, dynamic and activation of Nav1.7. Int J Biol Macromol 278, 134219 (2024).
Anil K. Kalia et al., Hyperexcitability of stem cell-derived sensory neurons of erythromelalgia patient normalized by divalent ions, in submission, 2024.
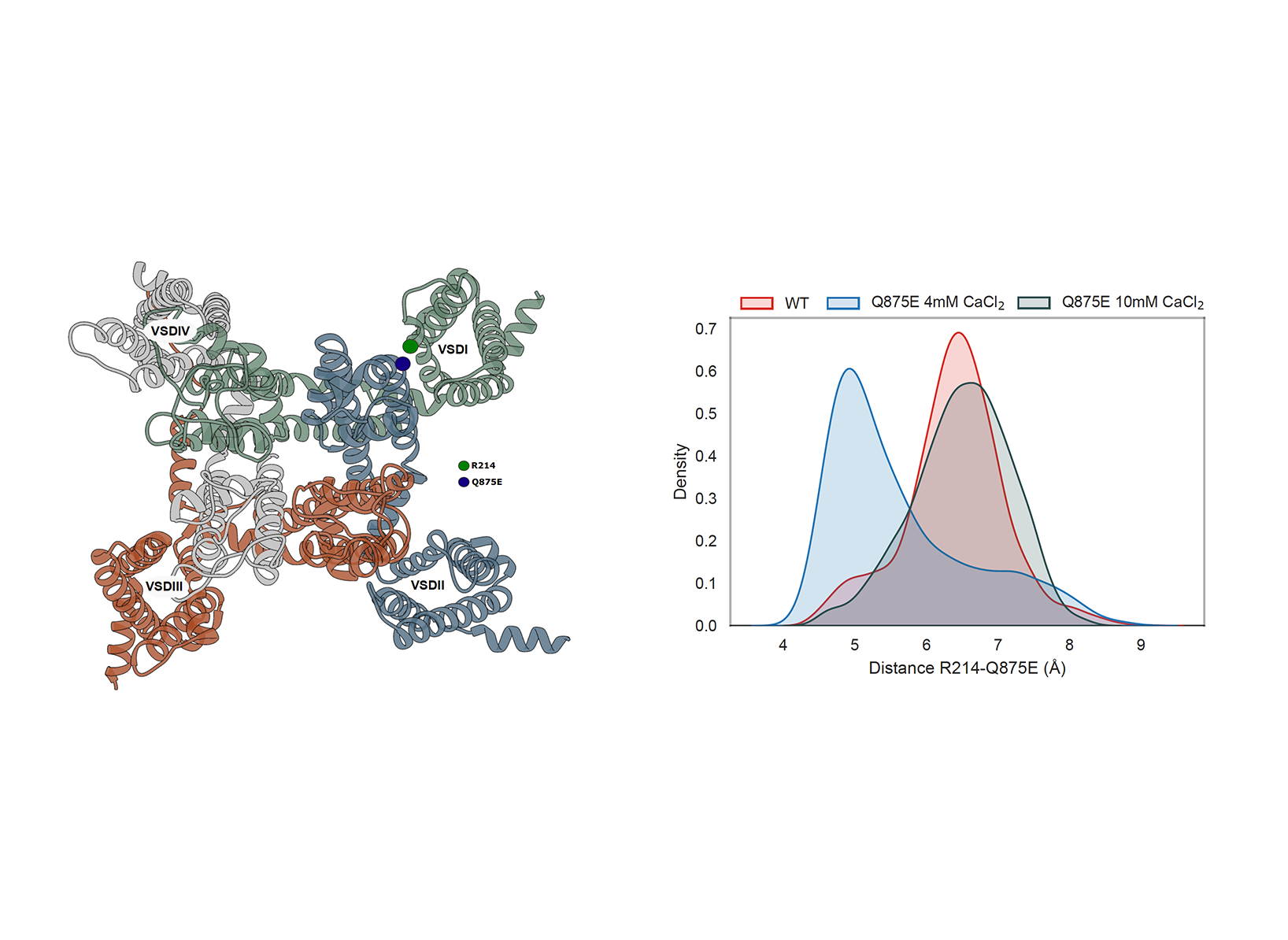 Figure 1: Left: Extracellular view of the channel; Domain I-IV in green, blue, red,
grey; Highlight on residues R214 on Domain I (green circle) and Q875E (blue
circle) on Domain II ; Visualized as ribbon in Chimera. Right: Distribution
of the distance between residues R214 and Q875E along the simulation in the
following systems: WT (red), Q875E mutant at control calcium concentration
(blue), Q875E mutant at increased calcium concentration (grey).
Figure 1: Left: Extracellular view of the channel; Domain I-IV in green, blue, red,
grey; Highlight on residues R214 on Domain I (green circle) and Q875E (blue
circle) on Domain II ; Visualized as ribbon in Chimera. Right: Distribution
of the distance between residues R214 and Q875E along the simulation in the
following systems: WT (red), Q875E mutant at control calcium concentration
(blue), Q875E mutant at increased calcium concentration (grey).